Phatnani Lab
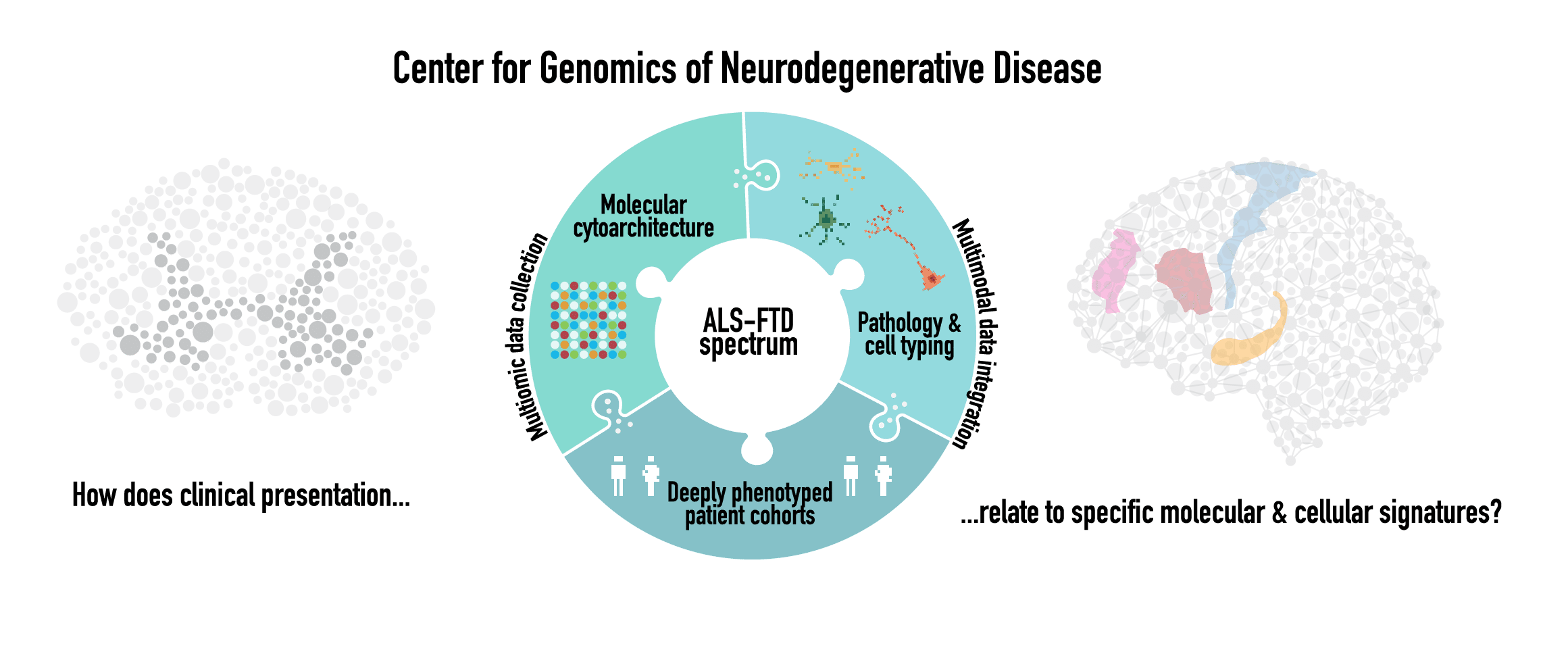
The vision of the Center for Genomics of Neurodegenerative Disease (CGND) is to establish a center for applying state of the art genetics, genomics, proteomics, and bioinformatics to the study of neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease.
Our Approach
To gain insights into the causes of neurodegenerative disease, the Phatnani lab examines the relationship between mutations, gene expression and disease mechanisms using whole genome sequencing, spatial transcriptomics, single nucleus (sn)-resolved epigenomics and snRNA-seq, and multiplex protein imaging. We apply these tools to both clinically well-characterized post-mortem human brain tissue of patients with ALS and mouse models of human disease to investigate the specific molecular underpinnings of functional impairment. Our ultimate goal is to unveil how disease-associated pathology is linked to dysfunction in specific cell types, which may in turn foster precision medicine-based treatment strategies.
Projects
Profiling Cellular and Molecular Dysfunction in the ALS-FTD Spectrum
- ALS and FTD are part of a neurodegenerative disease spectrum for which there are no biomarkers and limited treatments, stemming from a poor understanding of its pathogenesis. ALS is clinically heterogeneous with respect to a) whether motor symptoms are accompanied by cognitive impairment such as FTD, b) the site of onset of motor symptoms, c) the type of pathology observed in the brain and spinal cord, and d) which cell types are affected by this pathology. The sources of this heterogeneity are presently not well understood.
- We aim to create a transcriptomic and proteomic map with cellular and subcellular resolution of the brain and spinal cord of patients with ALS and ALS-FTD. This will allow us to identify pathological signatures, perturbations in intercellular interactions, and changes in gene expression correlated with site of symptom onset, and whether or not ALS is accompanied by FTD. To accomplish these aims, we use highly multiplexed protein imaging to deeply phenotype pathological changes in tissues and spatially resolved transcriptomic profiling to discover changes in gene expression profiles relative to these pathological features.
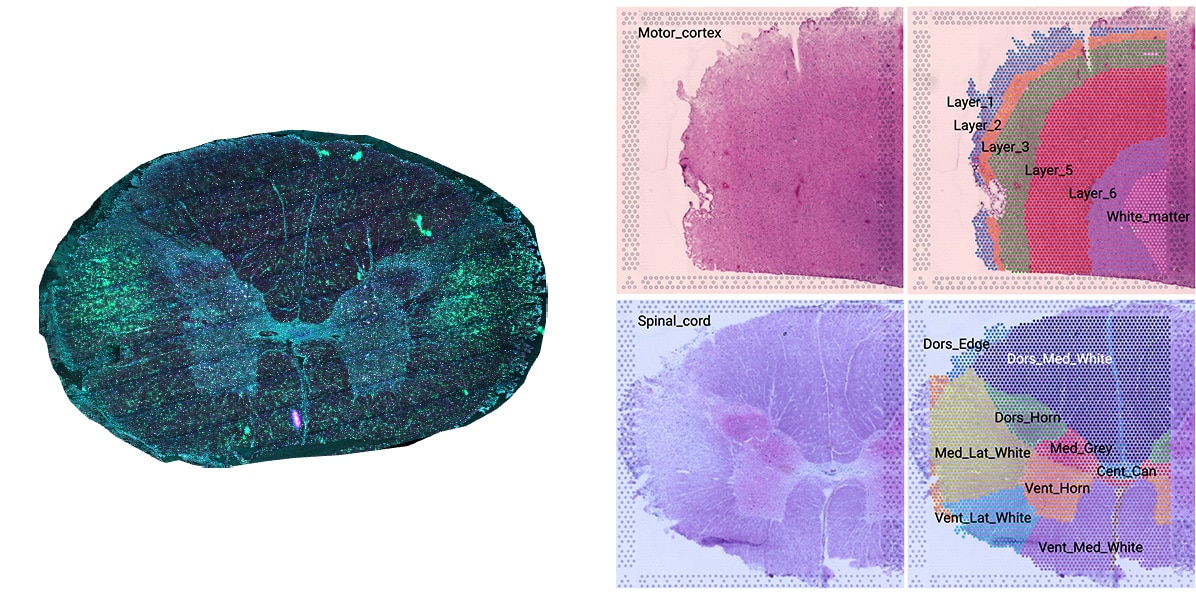
Left image: Multiplex human spinal cord staining of astrocytes, dendrites, macrophages and nuclei. Right image: Spatial transcriptomic gene expression and regional annotation for human brain and spinal cord.
Uncovering the Molecular Signatures of Cellular Senescence
- One of the adaptations cells make in an aging organism to maintain proper function is cellular senescence: the permanent arrest of cell division. Senescent cells are characterized by the expression of tumor suppressive genes, resistance to apoptosis, and a proinflammatory senescence associated secretory phenotype. By studying this small population of cells we hope to achieve a better understanding of age-related tissue dysfunction that leads to increased susceptibility to disease.
- To this end, our lab makes use of our robust spatial transcriptomics pipeline, paired with sn-RNAseq to characterize the heterogeneity of how cellular senescence presents at the single cell level within different CNS cell types. Using 4i, a highly multiplexed proteomic imaging platform, we’re additionally able to visualize up to 40 senescence biomarkers with subcellular resolution.
- In partnership with the University of Edinburgh Sudden Death Brain Bank and Columbia’s New York Brain Bank, we are accumulating non-pathological human spinal cord, dorsolateral prefrontal cortex, and hippocampus tissue with the ultimate goal is to develop a 3D multi-omic map of senescence across the adult aging spectrum.
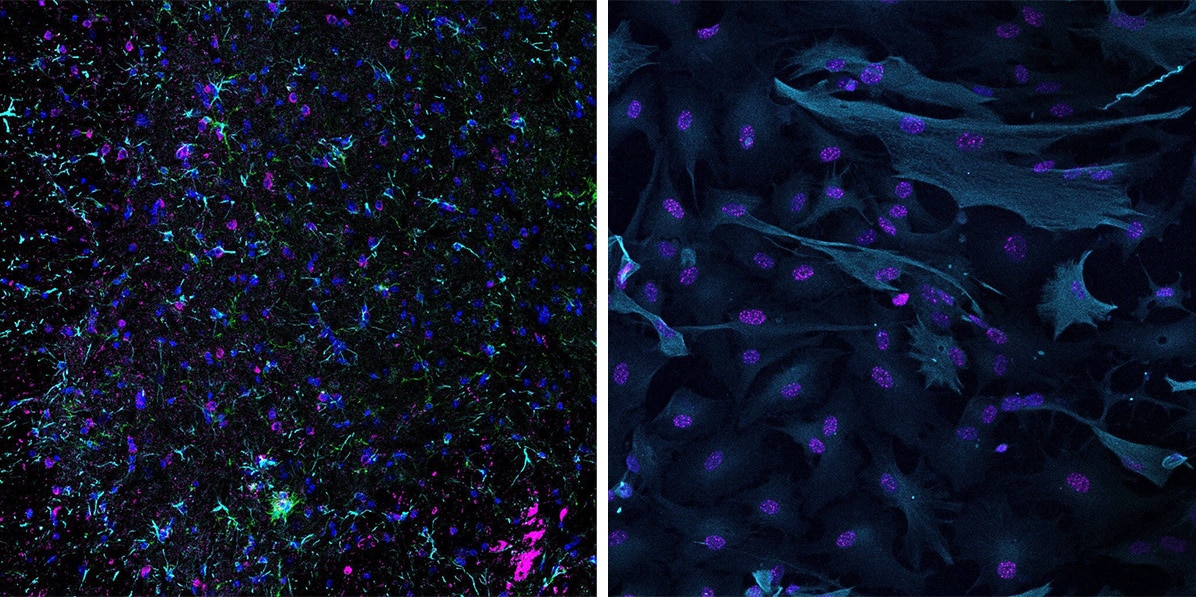
Left image: Microglia (green), astrocytes (cyan) and senescent cells (pink) in the ventral horn of the aged mouse spinal cord. Right image: Cultured primary human astrocytes induced for senescence using etoposide stained for astrocytes (cyan) and DNA damage (pink).
Hemali Phatnani, PhD
Director, Center for Genomics of Neurodegenerative Disease, NYGC
Assistant Professor of Neurological Sciences (in Neurology), Columbia University

Latest News & Publications
-
Cell Reports · March 10, 2025
-
bioRxiv · February 27, 2025 · Preprint
Differential Cellular Mechanisms Underlie Language and Executive Decline in Amyotrophic Lateral Sclerosis.
-
bioRxiv · February 20, 2025 · Preprint
Uncovering the Signatures of Cellular Senescence in the Human Dorsolateral Prefrontal Cortex.
The Consortium Undergraduate Student Program (CUSP)
The Consortium Undergraduate Student Program (CUSP) is a summer research internship program for undergraduate students hosted by laboratories participating in the NIH Common Fund’s Cellular Senescence (SenNet) program. We welcome applications from students of populations underrepresented in the clinical and biological sciences.
Center for Genomics of Neurodegenerative Disease Team

Postdoc and Graduate Students
-

Güney Akbalik
Postdoctoral Research Associate
-

Alexandria Awai
Visiting Scientist
-

Natalie Barretto
PhD Student
-

Zoe Butti
Postdoctoral Research Associate
-

Mark De Los Santos
PhD Student
-

Ben Fullerton
MD-PhD Student
-

Brhan Gebremedhin
Post-Baccalaureate Fellow
-

Risako Kimura
PhD Student
-

Olena Kuksenko
PhD Student
-

Nick Sloan
PhD Student
-

Samantha Zaninelli
PhD Student
Research Scientists
-

Lilian Coie
Staff Scientist
-

Aidan Daly
Computational Scientist
-

Shaunice Grier
Research Worker
-

Kennedy Harris
Lab Technician
-

Christopher Jackson
Senior Computational Scientist
-

Anurag Katiyar
Research Technician
-

Imdadul Haq
Associate Research Scientist
-

Luke Reilly
Associate Research Scientist III
-

Cláudio Gouveia Roque
Senior Staff Scientist
-

Patricia Santos
Research Assistant
Administration
-

Maria Hauge Pedersen
Scientific Program Manager
-

Mariecia Pook
Senior Program Manager
Alumni
-

Rawan Abdelaal
-

Cat Braine
-

Jan Bergmann
-

Obadele Casel
-

Demetra Catalano
-

Matthew Smith-Erb
-

Delphine Fagegaltier
-

James Gregory
-

Emily Hoelzli
-

Isabel Hubbard
-

Duyang Kim
-

Kristy Kang
-

Ian Laster
-

Matthew Leung
-

Kenneth Li
-

Silas Maniatis
-

Ariel Shepley-McTaggart
-

Joanna Petrescu
-

Nadia Propp
Contact Us

