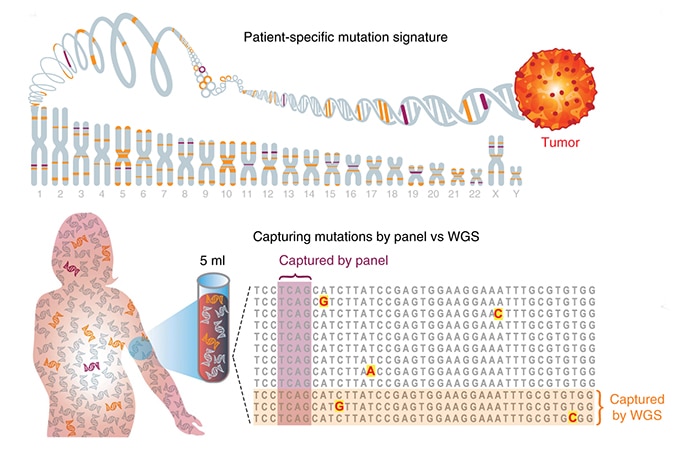
A DNA-sequencing strategy powered by machine learning may pick up even very low levels of tumor DNA in blood samples, potentially enabling the early detection of cancer recurrence after surgery or other treatments, according to a study led by scientists at Weill Cornell Medicine and the New York Genome Center (NYGC).
The scientists, who describe the new method in a study published on June 1 in Nature Medicine, demonstrated its potential value with blood tests on patients who had melanomas, colorectal cancers and lung cancers. The results showed that the new technique can be used to detect blood-borne tumor DNA even when it is much too sparse to be detected by existing methods.
“For many tumors the chance of recurrence after initial treatment is quite high, but up to now we haven’t always had a good way to detect those recurrences early, when they are most treatable,” said senior author Dr. Dan Landau, an assistant professor of medicine in the Division of Hematology and Medical Oncology at Weill Cornell Medicine, a core member of the NYGC, and an oncologist at NewYork-Presbyterian/Weill Cornell Medical Center.
“Liquid biopsy” methods employing gene-sequencing technology are already used to detect fragments of DNA that have come from tumor cells and are circulating in the bloodstream. However, these methods, while they are highly sensitive in principle, target a handful of specific genes or hotspots in the genome where cancer-associated mutations are expected. As Dr. Landau and colleagues noted in their analysis, targeted sequencing approaches can work only when circulating tumor DNA in blood samples includes sufficient fragments containing the targeted mutations—yet in many cases, for example following surgery to remove a tumor, the “tumor burden” is much too low for that. In fact, standard targeted-sequencing techniques often miss residual DNA from small tumors that are evident on standard imaging scans.
“It occurred to us that in some cases, it would be better not to look at a few DNA hotspots very thoroughly, but instead to make the most of available tumor DNA by detecting mutations across the entire genome,” said Dr. Landau, who is also a member of the Sandra and Edward Meyer Cancer Center at Weill Cornell Medicine.
The new method therefore involves the sequencing of all circulating DNA in a patient’s blood sample—a “whole genome” approach that boosts the effective sensitivity by orders of magnitude. The resulting DNA sequence data are evaluated by a machine-learning algorithm that looks for matches, across the genome, between mutations in the blood-borne DNA and any of the thousands of mutations detected in the prior sequencing of the patient’s primary tumor. The technique is sensitive not just to individual mutations in DNA but also to larger abnormalities in which segments of the genome have been deleted or multiplied.
Dr. Landau and colleagues performed proof-of-principle tests of the new method with melanoma patients, and found that it can accurately track their tumor burden as that burden changes during immunotherapy. The scientists then showed that the method can detect tumor DNA in some lung cancer and colorectal cancer patients following surgery—a very low tumor burden condition in which standard targeted sequencing methods are often limited in the ability to detect residual cancer. The detection of residual cancer in these patients was associated with worse long-term outcomes.
Dr. Landau and his colleagues hope that the new method ultimately will enter routine use for the early detection of recurring tumors and the monitoring of tumor burdens more generally.
“In most areas of medicine, treatments are guided by ongoing disease monitoring with blood tests or other methods,” Dr. Landau said. “Solid tumor oncology has been an exception, but we’re hoping that our new technology will help close that gap.”
He and his colleagues are continuing to develop their technique to make it even more sensitive and specific, and are hoping to confirm its utility in guiding cancer care in the context of clinical trials.
Dr. Landau has founded a company called C2i Genomics with co-first author Dr. Asaf Zviran, a member of his laboratory during the study, and others to commercialize the new technique. Co-first author and lab member during the study, Dr. Rafael Schulman, is also employed at the company. Dr. Landau has also participated in an advisory board for Illumina, Inc.
Read related news:
C2i, A Genomics SaaS Product to Detect Traces of Cancer, Raises $100M Series B
(Tech Crunch, April 15, 2021)
C2i Genomics Raises $12 Million in Series A Financing
(Business Wire, June 9, 2020)
Liquid Biopsy Startup C2i Genomics to Launch International Validation
Trials for MRD Detection Tech (GenomeWeb, June 12, 2020)
C2i Genomics Raises $12M Series A to Bring Whole Genome Pattern Recognition to Liquid Biopsy
(BioWorld, June 12, 2020)